One of the main mechanisms of genetic change involves enzymes called recombinases which catalyse the rearrangement of DNA sequences by breaking the DNA and rejoining the ends in new ways. This cutting and recombination is responsible for genetic diversity arising in sexual reproduction, and is also crucial for DNA repair and other cellular processes. Recombinant techniques are, of course, a key component in genetic engineering.
Despite their significance, little is known about structural changes that happen during the process. With this in mind, a team at the University of Glasgow have been studying the interaction of Tn3 resolvase with DNA. This recombinase binds with two specific sites on DNA, then breaks and recombines them. The catalytic process involves the formation of a transient structural assembly called a synaptic complex, which contains twelve copies of the resolvase and two pieces of the DNA sequence.
The researchers chose to work on a laboratory-made, highly-active variant of Tn3. This resolvase readily forms a simple, stripped-down version of the synaptic complex called an X-synapse, which is stable enough to be studied. It consists of four resolvase units bound to two short pieces of double-stranded DNA, and provides a useful structural model for investigating the process at the molecular level.
Now you see it
Small angle scattering, with both X-rays and neutrons, was used to analyse the synapse structure in its natural watery environment. Neutrons are particularly useful because the neutron scattering strength of the water can be matched to that of either the enzyme or DNA component by substituting a proportion of the water with heavy water (D2O). This makes that component ‘invisible’ in the neutron scattering pattern, so that only the other component is seen. In this way, the research team was able to ascertain the relative positions and conformation of the resolvase enzyme and DNA molecules by comparing the results with those predicted from computer models.
Models had indicated that the two DNA molecules, when brought together and tethered by Tn3 resolvase, would be kinked. The question was whether the DNAs would be kinked outwards and located on the outside of the complex (as in figures a and b), or bent inwards and on the inside (as in figure c). The neutron measurements showed that the DNAs bent outwards and sat on the outside of the X-synapse. This architecture has important implications because it keeps the sites of DNA cleavage far apart so it is not clear how the breaking and recombination happens. Clearly there is still a lot to learn.
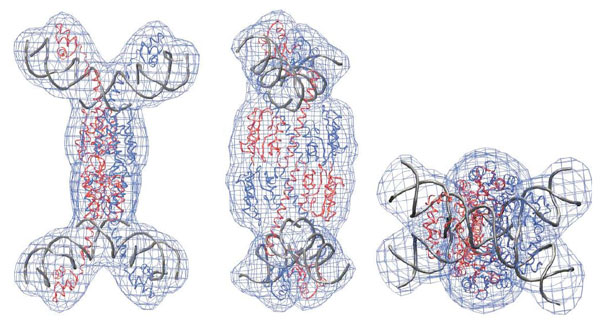
Mesh representation of three views of the final model for the X-synapse solution structure
View full-size image
Further Information
Solution Structure of the Tn3 Resolvase-Crossover Site Synaptic Complex, M Nöllmann et al, Molecular Cell 16 (2004) 127.
Behavior
of Tn3 Resolvase in Solution and its Interaction with res, M Nöllmann, O
Byron and W Marshall Stark, Biophysical Journal 89 (2005) 1920.