Taste is an important sense and has many functions including identification of energy-rich nutrients, protection against the intake of potentially poisonous chemicals as well as contributing to the overall enjoyment of eating. The main flavour components tasted by humans are salty, sweet, bitter, sour and umami but the complexity of tasting means that we have the ability to distinguish many different flavours. However the process of tasting is very complicated and poorly understood. An increased understanding of the mechanism of tasting could be applied to areas such as the design of food components which can be tailored to be healthier without sacrificing flavour.
Tastes are sensed by multiple receptor proteins; sweet and bitter tastes are especially associated with T1R2, T1R3 and T2R receptors. Molecules with a complementary shape to these receptor proteins bind to them causing a message to be sent to the brain which triggers the release of neurotransmitters resulting in a sweet or bitter taste. Sugar molecules bind to receptors at multiple sites and how many of these sites a molecule interacts with is thought to affect how sweet a molecule tastes. Binding affinity may also be a contributing factor.
Scientists from Università di Roma Tre and the University of Oxford have been using the SANDALS diffractometer at ISIS Neutron and Muon Source to investigate the hydration of sugar molecules using neutron diffraction. Glucose and mannose were studied as they are two sugar molecules with different degrees of sweetness. In water, each sugar exists in 2 structural forms (α and β anomers). Glucose is tasted as being sweeter than mannose, and while α-mannose is sweet, the β-form can elicit a bitter aftertaste. It thus remains an intriguing question as to what determines a sugar to be perceived sweeter than another, given the relatively small differences between the stereochemistry of sugars.
Data collected from this experiment on the hydration of mannose was studied alongside previously recorded data on the structure of hydration in glucose (Maugeri et al. 2017. Structure-activity relationships in carbohydrates revealed by their hydration. Biochimica et Biophysica Acta (BBA)-General Subjects, 1861(6), 1486-1493.). Standard glucose and mannose and deuterated glucose and mannose (where each –OH hydrogen is deuterated) were used in different ratios allowing for a more accurate view of the atoms. While the structure of the receptors is still unknown, there are models for the receptor-sugar binding mechanism. By mapping the sites of hydrogen bonding on the sugar molecules using neutron diffraction, a comparison can be made to the receptor binding models to determine if there is a link between this and the perceived sweetness of each molecule. The extent of hydrogen bonding of the glucose and mannose anomers can also be compared.
|
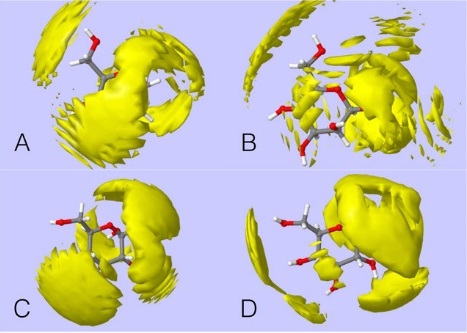
Figure 1. Shows the hydration around glucose and mannose molecules. A, B, C, D refer to α- and β- anomers of mannose and glucose, respectively. Reprinted and adapted with permission from Rhys. N et al, 2017, DOI: 10.1021/acs.jpcb.7b03919 . Copyright 2017
|
Neutron diffraction experiments on
SANDALS (Small Angle Neutron Diffraction for Liquid and Amorphous Sample) revealed that glucose and mannose form different numbers of hydrogen bonds with water, with glucose forming more than mannose. Both α and β anomers of glucose form strong hydrogen bonds with water, and a lthough the α-form of mannose bonds in a similar way to glucose, the β-form (which is reported to taste bitter) does not match the same model. The different structural forms also showed varying amounts of hydrogen bonding around O3-O4-O5 (fig 1) with α-glucose exhibiting the most and β-mannose having far fewer. β-mannose therefore has a smaller hydrogen bond surface area to the receptors and subsequently is likely to bind to the T1R2+T1R3 receptors with lower affinity. Furthermore, while glucose was found to interact with water as both an acceptor and donor when forming hydrogen bonds, mannose only acted as a donor to surrounding water molecules. This is likely to correspond to the binding affinity and therefore the sweetness of glucose compared to mannose.
“According to the "sweetness triangle model" proposed by Shallenberger and Acree (Shallenberger, R. S.; Acree, T. E. Molecular Theory of Sweet Taste. Nature 1967, 216, 480−482.) the molecular conformation and relative distance between the H donor/acceptor sites on the molecule is the requisite for eliciting the sweet taste. Our data, while confirming this assumption, suggest that the sweetness ranking is determined by the strength of the H bonding, as measured by the position of the sugar-water H-bond peak, with fructose and glucose (the sweetest monosaccharides) forming shorter H-bonds than mannose (sweet with bitter aftertaste) and the disaccharide trehalose (having no taste at all)."
Maria Antonietta Ricci, Università di Roma Tre
It is clear that further studies are necessary, however a link between position and strength of hydrogen bonds and receptor binding areas with degree of sweetness is evident. However why β-mannose tastes bitter cannot be deduced from these results and requires further investigation of the sugar's interaction with the T2R receptor which is known to be involved with tasting bitter. Further discoveries into how certain molecules are perceived as being sweeter could aid in the development of food components such as artificial sweeteners, enabling sweeter tasting sweeteners to be created that can be used in much smaller quantities.
Further information
The research paper can be found here Glucose and Mannose: A Link between Hydration and Sweetness, N. H. Rhys, F. Bruni, S. Imberti, S. E. McLain, and M. A. Ricci The Journal of Physical Chemistry B 2017 121 (33), 7771-7776 DOI: 10.1021/acs.jpcb.7b03919 .